A research team led by Dr Gary Ying Wai CHAN from the School of Biological Sciences at The University of Hong Kong (HKU), has uncovered a new mechanism that ensures correct DNA segregation in cell division, where improper cell division will lead to the development of cancer.
The team’s findings, published in the journal Cell Reports, focus on the roles of two proteins, RIF1 and protein phosphatase 1 (PP1), in resolving ultrafine DNA bridges. These bridges are formed when sister chromatids are connected by DNA joint molecules during mitosis. If these DNA bridges cannot be resolved or removed properly, they will eventually break and cause DNA damage in the daughter cells, which can lead to the development of cancer cells.
Highlights
- Depletion of RIF1 induces single-stranded ultrafine anaphase bridges (UFBs)
- Protein phosphatase 1 (PP1) is recruited to UFBs via RIF1
- CDK1 negative regulates RIF1-PP1 interaction via the CIII domain of RIF1
- Phosphorylation-resistant mutant of PICH bypasses the need of RIF1
The process of Mitosis
A human life begins with a single cell – the fertilised egg. This single cell needs to replicate and divide into approximately 37 trillion cells. The process by which a cell replicates its DNA then equally segregates into two identical cells is known as mitosis, which is a vital process for growth and replacing worn out cells. However, equal segregation of DNA is one of the most challenging tasks in mitosis.
Our DNA is organised into 23 pairs of chromosomes, each of which is replicated into two sister chromatids. During mitosis, these sister chromatids are separated into two identical daughter cells. They are often connected by DNA joint molecules, which can form long fine DNA threads known as ultrafine DNA bridges when the two sister chromatids are separating under a pulling force. If these DNA bridges cannot be resolved or removed properly, they will eventually break, causing damage in the daughter cells. In a worst-case scenario, this DNA damage can potentially lead to the development of cancer.
To elucidate the detailed mechanism of how cells resolve the DNA bridges, the team led by Dr Gary Ying Wai Chan uncovered the roles of two proteins, RIF1 and protein phosphatase 1 (PP1) in regulating the resolution of ultrafine DNA bridges.
The role of RIF1 and PP1 in resolving DNA bridges
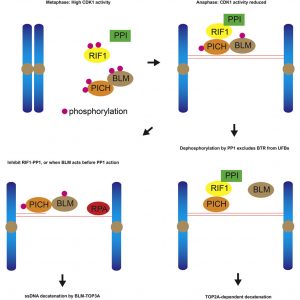
In 2007, two research groups led by Professor Erich NIGG and Professor Ian HICKSON discovered the existence of ultrafine DNA bridges by identifying the first two ultrafine bridge-binding proteins, PICH and BLM. Later on, RIF1 was also identified as a bridge-binding protein. It is known that PICH is the first protein recognising the DNA bridges, then it recruits BLM and RIF1 to ultrafine DNA bridges; however, the exact mechanism by which these bridge-binding proteins work to resolve the bridges remains unclear.
To understand the role of RIF1 in resolving DNA bridges, the research team employed a revolutionary genome editing technology, the CRISPR/Cas9, to deplete RIF1 in a cell model during mitosis. During which, the team found that loss of RIF1 leads to increased formation of DNA damage and micronuclei due to breakage of ultrafine DNA bridges, as our data suggest that RIF1 plays a crucial role in preventing double-stranded DNA bridges from converting into single-stranded DNA, which is more susceptible to breakage.
RIF1 was found to achieve this by recruiting PP1, a protein phosphatase, which reduces the interaction between BLM and PICH and thereby reduces the amount of BLM on the bridges, which the team discovered that is responsible for unwinding the bridge DNA to single-stranded DNA.
Finally, the double-stranded DNA bridges protected by RIF1-PP1 are believed to be resolved by another enzyme known as topoisomerase IIα, which could mediate double-stranded DNA decatenation to ensure proper DNA segregation.
This research not only sheds light on the novel regulatory mechanism by which RIF1-PP1 facilitates the resolution of DNA bridges, but also reveals how ultrafine DNA bridges can induce DNA damage and genome instability if they are not properly resolved.
The findings suggest that DNA bridge-binding proteins may serve as potential therapeutic targets for the development of anticancer drugs, as DNA bridges are considered a source of genome instability that drives tumorigenesis.
‘The discovery that RIF1 recruits PP1 to the bridges is the first step in understanding how the resolution of ultrafine DNA bridges is regulated by protein phosphorylation/dephosphorylation,’ said Dr Gary CHAN. ‘The next step is to identify the target substrates of the RIF1-PP1 complex, which can advance our understanding of how different bridge-binding proteins interact with each other and may lead to the identification of new therapeutic targets for cancer.’
Summary
Resolution of ultrafine anaphase bridges (UFBs) must be completed before cytokinesis to ensure sister-chromatid disjunction. RIF1 is involved in UFB resolution by a mechanism that is not yet clear. Here, we show that RIF1 functions in mitosis to inhibit the formation of 53BP1 nuclear bodies and micronuclei. Meanwhile, RIF1 localizes on PICH-coated double-stranded UFBs but not on RPA-coated single-stranded UFBs. Depletion of RIF1 leads to an elevated level of RPA-coated UFBs, in a BLM-dependent manner. RIF1 interacts with all three isoforms of protein phosphatase 1 (PP1) at its CI domain in anaphase when CDK1 activity declines. CDK1 negatively regulates RIF1-PP1 interaction via the CIII domain of RIF1. Importantly, depletion of PP1 phenocopies RIF1 depletion, and phosphorylation-resistant mutant of PICH shows reduced interaction with the BTR complex and bypasses the need of RIF1 in preventing the formation of single-stranded UFBs. Overall, our data show that PP1 is the effector of RIF1 in UFB resolution.
The journal paper, entitled ‘RIF1 Suppresses the Formation of Single-Stranded Ultrafine Anaphase Bridges via Protein Phosphatase 1’, can be found here
研究發現RIF1-PP1抑制 BLM解離DNA超細纖維橋
不當的細胞分裂有可能導致癌症的發生,因此科學家一直在尋找維持細胞分裂穩定的機制。香港大學生物科學學院助理教授陳英偉的研究團隊,成功發現了一種在細胞分裂時維持DNA分離穩定,防止「DNA橋」斷裂的新機制,有助更深入地了解細胞分裂過程,可望為開發抗癌藥物帶來新思路。
人類生命由受精卵經過不斷複製和分裂方能構成人體約37萬億個細胞,過程中細胞會將DNA複製並平均分配到兩個相同的子細胞,此過程稱為「有絲分裂」。在「有絲分裂」期間,細胞會形成連接姐妹染色單體的細長DNA分子,稱為「DNA橋」,而在分裂過程中一旦未能解離或解離不完全,最終會導致「DNA橋」斷裂並對子細胞中的DNA造成損傷,導致癌細胞的形成。
是次研究探討兩種蛋白質RIF1和蛋白磷酸酶1(PP1)在解離「DNA橋」中的作用,包括發現RIF1能抑制雙鏈「DNA橋」轉化為單鏈更易斷裂的「DNA橋」,意味着RIF1能保護細胞免受DNA損傷的侵害。
此外團隊亦發現,RIF1是通過募集PP1蛋白來實現此作用,揭示了RIF1-PP1促進「DNA橋」正常解離的新型調節機制。由於「DNA橋」斷裂被認為會導致基因組不穩定驅使腫瘤形成,是次研究亦說明,「DNA橋」結合蛋白可成為開發抗癌藥物的新潛在治療靶標。有關結果已於期刊《Cell Reports》發表。


